The reference genome of SARS-CoV-2 from the NCBI archive is required as the basis for all calculations.
This can be found via the following link:
The Fasta sequence can be downloaded via the "Send to" button. The gene annotation in GFF3 format is also required.
This will give you the following files:
- NC_045512.2.fasta
- NC_045512.2.gff3
The calculation of mutations can be performed for any genome from the central archive of GISAID.
However, this requires an account at GISAID.
In the case of the omicron variant, the sequences of interest must be selected and downloaded from the archive.
In the example shown, the entries with the currently occurring Omicron variant were selected and saved in multi-fasta format.
The name of the file for this example is 211129_B.1.1.519_GISAID.fasta.
It contains a total of 165 entries. A list of the used GISAID accession numbers can be found here.
A look at the file shows the following:
>hCoV-19/Hong_Kong/VM21044713-1/2021|EPI_ISL_6590782|2021-11-13
AGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGGCTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATT
AATAACTAATTACTGTCGTTGACAGGACACGAGTAACTCGTCTATCTTCTGCAGGCTGCTTACGGTTTCGTCCGTGTTGC
AGCCGATCATCAGCACATCTAGGTTTTGTCCGGGTGTGACCGAAAGGTAAGATGGAGAGCCTTGTCCCTGGTTTCAACGA...
>hCoV-19/Botswana/R40B60_BHP_3321001247/2021|EPI_ISL_6640917|2021-11-11
AGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGGCTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATT
AATAACTAATTACTGTCGTTGACAGGACACGAGTAACTCGTCTATCTTCTGCAGGCTGCTTACGGTTTCGTCCGTGTTGC
AGCCGATCATCAGCACATCTAGGTTTTGTCCGGGTGTGACCGAAAGGTAAGATGGAGAGCCTTGTCCCTGGTTTCAACGA
...
We can see that the entries start with the name of the sequence in the header line and
that the sequence information is stored in the following lines.
The sequences from GISAID are mapped to the reference genome using the minimap2 tool and are
then immediately written to a sorted BAM file.
minimap2 -ax asm5 NC_045512.2.fasta 211129_B.1.1.519_GISAID.fasta | samtools sort -o GISAID.sorted.bam
The mapped sequences are then used to determine the differences from the reference sequence.
From the mapped sequences, the deviations from the reference genome are determined and stored in a VCF (Variant call format).
After the index to the file has been created, we can assemble the consensus sequence using the bcftools tool.
The resulting consensus sequence now contains the specific mutations of the omicron variant in addition to the nucleotide sequences of the reference genome.
# Create read pileup and pipe results to the variant caller
bcftools mpileup -Ou -f NC_045512.2.fasta GISAID.sorted.bam | bcftools call -mv -Oz -o GISAID.call.vcf.gz
# Create VCF index
bcftools index GISAID.call.vcf.gz
# use reference genome to create consensus sequence from the obtained mutations
cat NC_045512.2.fasta | bcftools consensus GISAID.call.vcf.gz > consensus.fa
In the downloaded genome annotation (NC_045512.2.gff3), we see that the spike gene is annotated as follows:
NC_045512.2 RefSeq gene 21563 25384 . + . ID=gene-GU280_gp02;Dbxref=GeneID:43740568;Name=S;gbkey=Gene;gene=S;gene_biotype=protein_coding;gene_synonym=spike glycoprotein;locus_tag=GU280_gp02
We use this information of the start and end coordinates to cut the sequence from the reference genome.
# use start and end coordinates to extract spike sequence
samtools faidx NC_045512.2.fasta NC_045512.2:21563-25384 > spike.fasta
The extracted spike sequence is then mapped to the previously created consensus sequence.
This gives us the positions of the spike gene on the assembled consensus sequence.
# map spike sequence to created consensus sequence
minimap2 -ax asm5 consensus.fa spike.fasta | samtools sort -o spike.sorted.bam
From the mapping, we now extract the positions of the spike gene in the consensus sequence. For this we use bedtools.
The second and third columns contain the start and end positions of the spike gene.
# convert from BAM to BED format
bedtools bamtobed -i spike.sorted.bam
NC_045512.2 21550 25363 NC_045512.2:21563-25384 60 +
Using the determined positions of the spike gene on the consensus sequence, we can extract the spike sequence of the omicron variant.
# extract spike sequence from obtained coordinates
samtools faidx consensus.fa NC_045512.2:21551-25363 > spike_omicron.fasta
## Replace fasta header from '>NC_045512.2:21551-25363' to '>spike_omicron' for better readability
sed -i -E 's/>.*/>spike_omicron/' spike_omicron.fasta
This gives us the spike sequence of the omicron variant.
The generated spike sequence and the GISAID sequence are mapped back to the reference genome to verify the correct incorporation of the mutations.
minimap2 -ax asm5 NC_045512.2.fasta spike_omicron.fasta | samtools sort -o spike_omikron.mapped.sorted.bam
minimap2 -ax asm5 NC_045512.2.fasta 211129_B.1.1.519_GISAID.fasta | samtools sort -o GISAID.mapped.sorted.bam
samtools index spike_omikron.mapped.sorted.bam
samtools index GISAID.mapped.sorted.bam
The BAM files are loaded into the IGV Genome Browser.
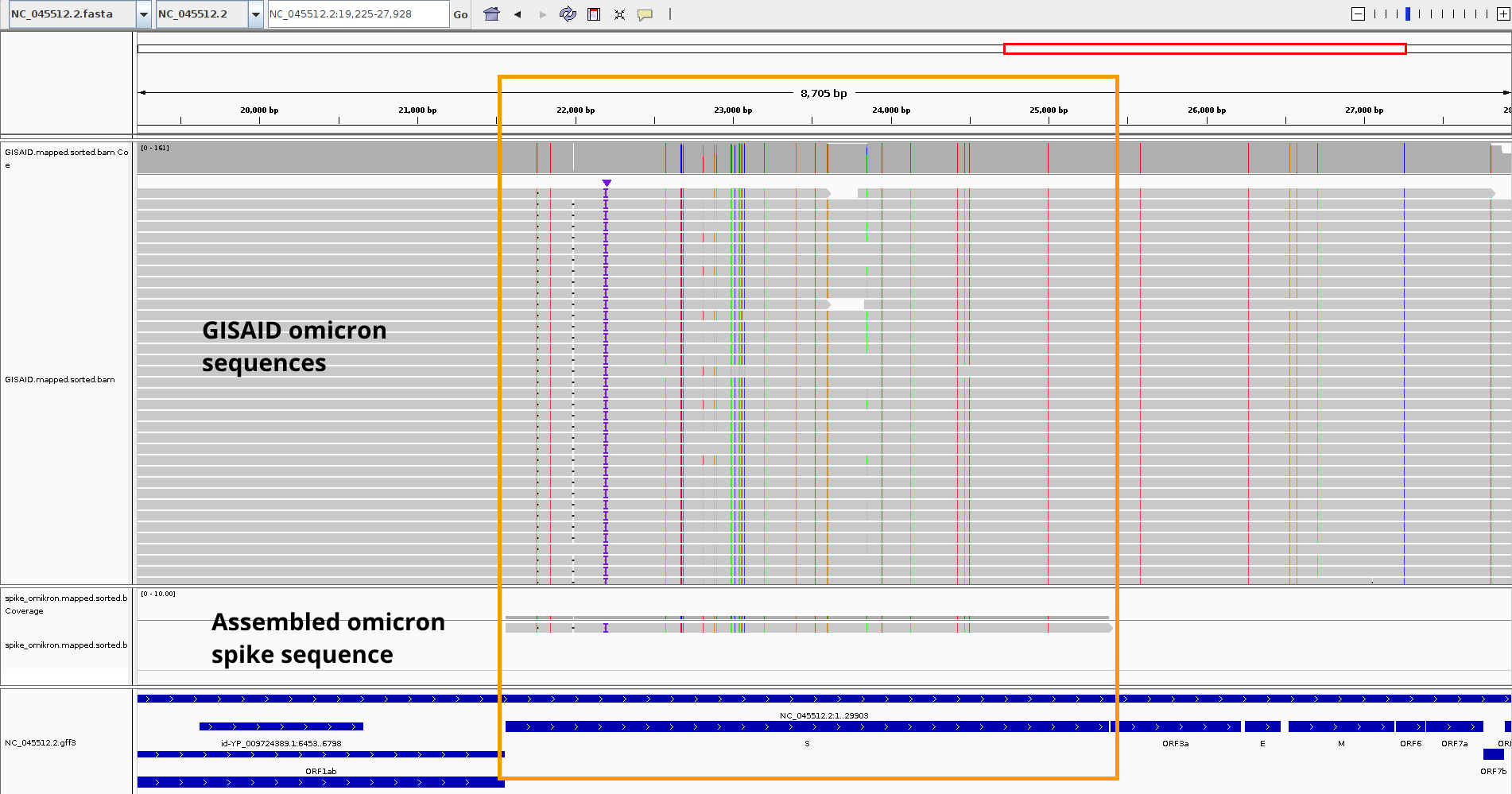
Our assembled spike sequence mapped to the position of the annotated
position of the spike gene in the reference genome and the specific mutations were integrated into the consensus sequence.
We now translate the created nucleotide sequence of the spike protein to amino acids. For this we use the tool transeq of the Emboss Package.
# translation of the omicron spike sequence
transeq -sequence spike_omicron.fasta -outseq spike_omicron_aminoAcid.fa
# translation of the reference genome spike sequence
transeq -sequence spike.fasta -outseq spike_referenceGenome_aminoAcid.fa
We compare the omicron amino acid sequence with the reference amino acid sequence to see the differences at that level as well.
# Merge spike sequences from reference and omikron lineage
cat spike_referenceGenome_aminoAcid.fa spike_omicron_aminoAcid.fa > all_spikes.fa
# make a multiple alignment from both spike sequences
mafft all_spikes.fa > all_spikes.mafft.fa
The MAFFT alignment of the amino acid sequences of the spike protein is visualized in Jalview.
